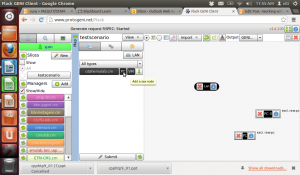
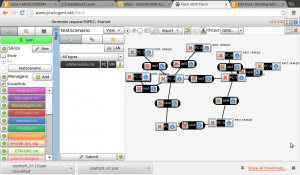
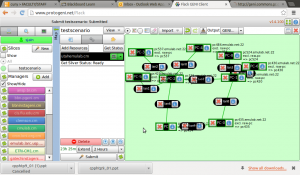
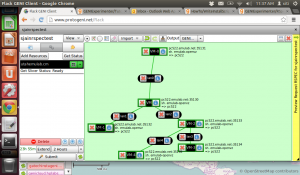
Lets run a quick sample experiment to see how this whole thing ties in together. I will do this the hard way i.e., manually just to test the waters and understand what I need for an automated experiment.
I have constructed a network graph and I am logging on to one of the machines (ssh) to run the iperf server.
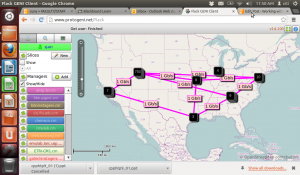
- Sample network graph
I will start an iperf server on VM-0 and an iperf client on VM-4.
Note that I will need to use sudo with all commands that I type here because al commands run as a user with super user priviledge.
When I looked for iperf command on VM-0, I got an error, see below
[user@VM-0 ~] sudo which iperf
iperf: Command not found
This tells me that iperf is not installed, so make a note that I need to install iperf before I can start my experiment. Since this machine is running fedora, I will use the yum package manager to install iperf
[user@VM-0 ~] sudo yum -y install iperf
I am interested in the last few lines of the output
Installed:
iperf.x86_64 0:2.0.5-3.fc15
Lets check where iperf is located now:
[user@VM-0 ~]$ sudo which iperf
/usr/bin/iperf
Note that the PATH variable is not necessarily set, so we need to provide the entire path to commands unless we explicitly set the PATH environment variable.
Now I start my iperf server
[user@VM-0 ~]$ sudo /usr/bin/iperf -s -p 5001 > iperf_server.txt
————————————————————
Server listening on TCP port 5001
TCP window size: 85.3 KByte (default)
————————————————————
Next I will follow the same steps and run the iperf client on VM-4.
[user@VM-4 ~]$ sudo /usr/bin/iperf -c VM-0 -p 5001 -t 90 -i 1 > iperf_client.txt
————————————————————
Client connecting to VM-0, TCP port 5001
TCP window size: 16.0 KByte (default)
————————————————————
[ 3] local 10.10.5.2 port 54339 connected with 10.10.1.1 port 5001
[ ID] Interval Transfer Bandwidth
[ 3] 0.0- 1.0 sec 11.9 MBytes 99.6 Mbits/sec
[ 3] 1.0- 2.0 sec 11.4 MBytes 95.4 Mbits/sec
[ 3] 2.0- 3.0 sec 11.5 MBytes 96.5 Mbits/sec
[ 3] 3.0- 4.0 sec 11.5 MBytes 96.5 Mbits/sec
[ 3] 4.0- 5.0 sec 11.2 MBytes 94.4 Mbits/sec
[ 3] 5.0- 6.0 sec 11.5 MBytes 96.5 Mbits/sec
Once this experiment finishes, I can collect my data from the files that I stored it in i.e.
iperf_client.txt and iperf_server.txt